Poisson random number generation
In the birdnest simulator, I use Poisson random numbers a great deal. In fact, given n individuals, each generation t must generate n Poisson random numbers. This is exceeded only by the number of uniform random numbers, for which I use TinyMT. The simplest Poisson generator is Knuth's method:
size_t
poisson_small(void *mt, float l)
{
float L, p;
size_t k;
L = expf(-l);
k = 0;
p = 1.0;
do {
k++;
p *= random_unit(mt);
} while (p > L);
return(k - 1);
}
Unforunately, this scales linearly with mean λ. Since the simulation model is intended to work with λ → ∞, and even in practise one sees λ > 30, this is unacceptable. I then ported John Cook's implementation of Atkinson's rejection method, built for values λ > 30.
size_t
poisson_large(void *mt, float l)
{
const double c = 0.767 - 3.36 / l,
beta = M_PI / sqrtf(3.0 * l),
alpha = beta * l,
k = logf(c) - l - logf(beta);
float u, x, v, y, temp, lhs, rhs;
int n;
for (;;) {
u = random_unit(mt);
x = (alpha - logf((1.0 - u)/u))/beta;
n = (int)floorf(x + 0.5);
if (n < 0)
continue;
v = random_unit(mt);
y = alpha - beta * x;
temp = 1.0 + expf(y);
lhs = y + logf(v/(temp*temp));
rhs = k + n*logf(l) - log_factorial(n);
if (lhs <= rhs)
return(n);
}
}
While faster than Knuth for large values of λ, sublinear performance gains were met at λ
high enough to be unreasonably costly to compute.
In other words, although faster, the wall-clock time of computing such high values of λ makes it unacceptable.
I use Cook's Stirling's approximation to compute log_factorial().
Solutions
The simulator computes Poisson means from a payoff matrix, so one can explicitly bound values of λ from the maximum of this matrix. Since I know I have a fixed bound, and that Poisson means are natural numbers, I can precompute distributions from the finite set of possible means. To do so, I use the Poisson probability mass function (PMF), (λke-λ)/k!.
struct cache {
float l;
size_t bucketsz;
size_t buckets[CACHE_BUCKETS];
};
size_t
poisson_cached(void *mt, float l)
{
static struct cache *c;
uint32_t key, hash;
size_t mmax, i, j, k;
float v, a, b;
if (0.0 == l)
return(0);
if (NULL == c)
c = xcalloc(CACHE_SIZE,sizeof(struct cache));
hash = (uint32_t)l;
key = hash % CACHE_SIZE;
if (l != c[key].l) {
c[key].l = l;
k = i = 0;
while (k < CACHE_BUCKETS) {
a = i * logf(l);
b = -l;
v = a + b - log_factorial(i);
mmax = expf(v) * CACHE_BUCKETS;
for (j = 0; j < mmax; j++, k++)
c[key].buckets[k] = i;
if (i++ > hash && 0 == mmax)
break;
}
c[key].bucketsz = k;
}
return(c[key].buckets[random_uint
(mt, c[key].bucketsz)]);
}
I fill m buckets with m Pr(X = x) of a given number x.
To compute a Poisson number, one accesses bucket mx, where x is a uniform random
number in the unit interval.
I compute the natural logarithm of the PMF, and thus the log factorial, instead of doing so directly to avoid computing
factorials.
I can then reuse Cook's efficient log_factorial() function.
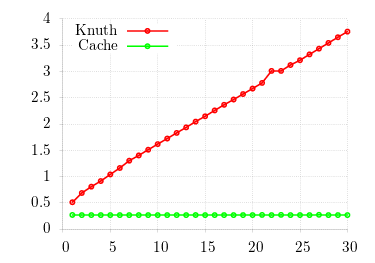
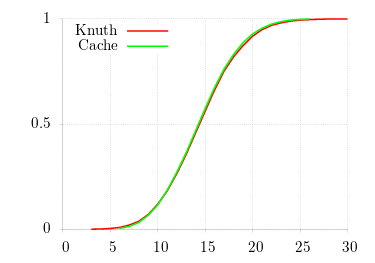
For correctness, I fix λ = 15 and show the CDF (using gnuplot's cumulative smoothing function) for 104 samples. One can see the constant-time access of the cache algorithm as well as the accuracy of its results.